COVID19: What is it and How is it Spread?

By Najung Lee (Pembroke) & Ragavi Vijayakumar (Downing). April 6, 2020.
COVID-19 and SARS-CoV-2
Coronavirus Disease 2019 (COVID-19) is an infectious disease caused by SARS-CoV-2 virus. It was first identified in Wuhan, capital of Hubei province of China and has resulted in a global pandemic.SARS-CoV-2 is a type of coronavirus (CoV), which is a family of enveloped positive-stranded RNA virus that infects vertebrates1. Specifically, it is a strain of severe acute respiratory syndrome-related coronavirus (SARSr-CoV) – a type of coronavirus that enters the host cell by binding to angiotensin-converting enzyme 2 (ACE2) receptor on cell membranes2. These receptors are transmembrane proteins found on lungs, arteries, heart, kidney and intestinal cells.
Bats are considered to be the reservoir of many strains of SARSr-CoV. Genetic analysis of the viral genome of SARS-CoV-2 revealed a high level of similarity to bat coronavirus, suggesting that SARS-CoV-2 is a bat-borne coronavirus3. Next-generation sequencing analysis of bronchoalveolar lavage fluid (BALF) samples from patients in Wuhan, revealed that 87.1% of the sequences matched the sequence of bat-borne SARSr-CoV. Many of SARSr-CoV strains are known to be non-human infecting species.
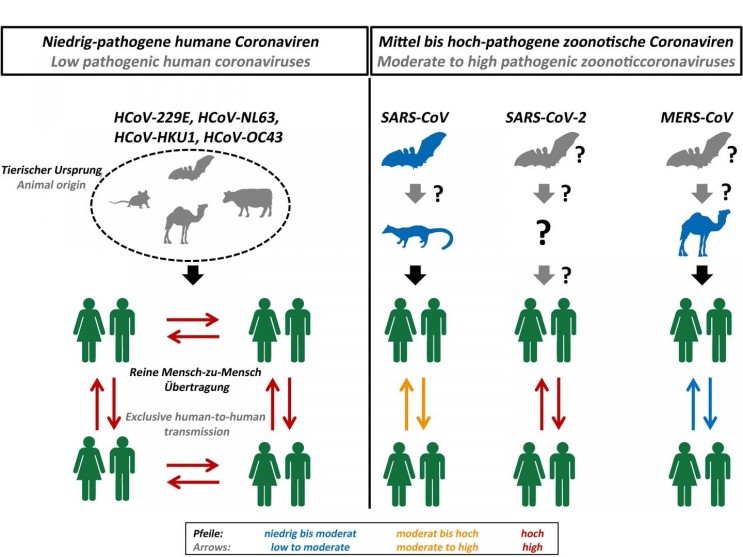
Figure 1. Origin and transmission of pathogenic coronaviruses. Credit: Markus Hoffmann
How does SARS-CoV-2 infect humans?
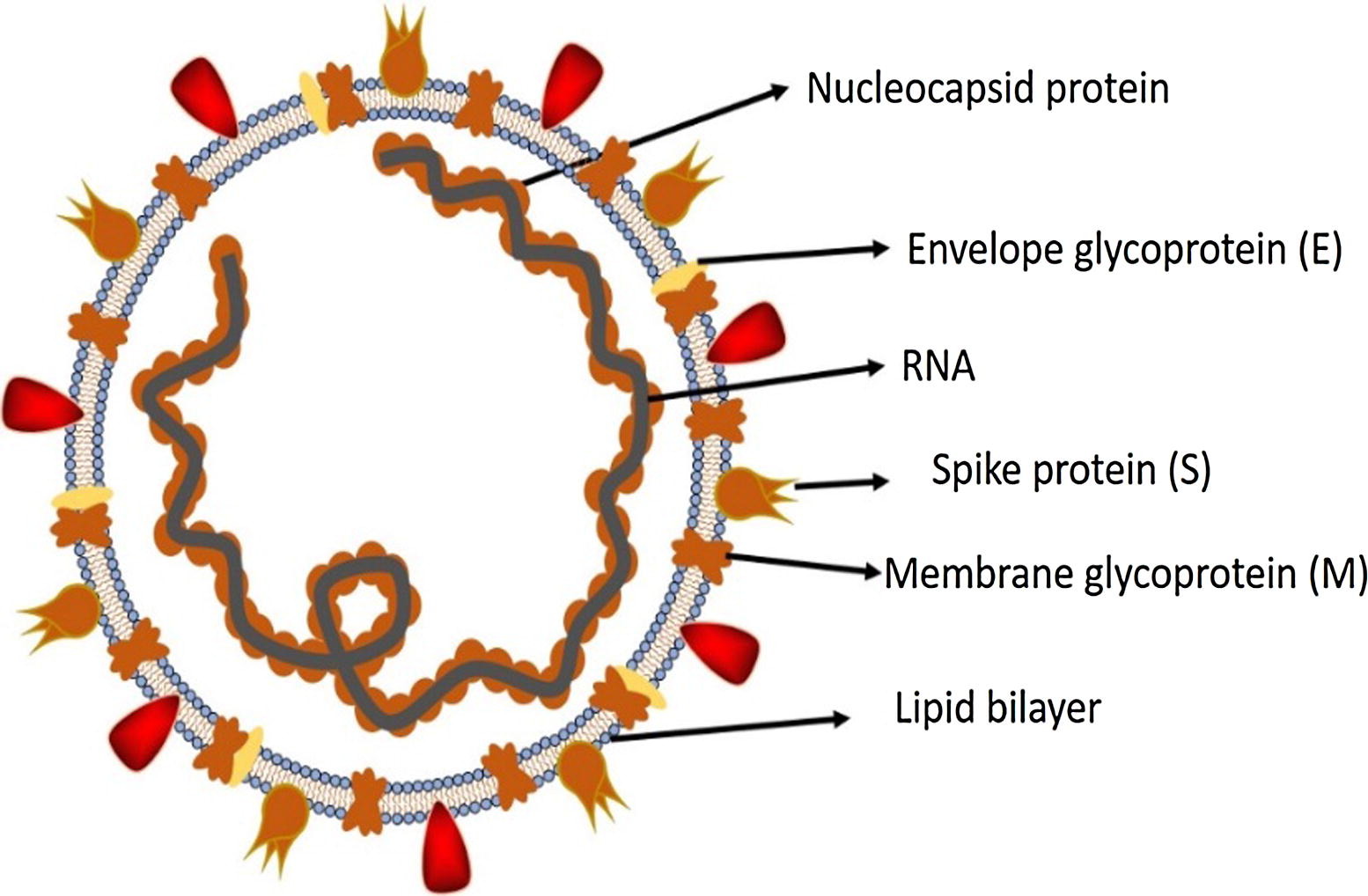
Figure 2. Structure of SARS-CoV-2. Credit: https://doi.org/10.1016/j.jare.2020.03.005
The S protein is a trimeric fusion protein and undergoes structural changes between up and down conformation to promote fusion of the viral membrane and the host cell membrane4. One of the RBD is used for the initial attachment to the ACE2 receptor. The structure of the RBD showed great similarity to the one of SARS-CoV, therefore it has been (re)named from 2019-nCoV to SARS-CoV-2.
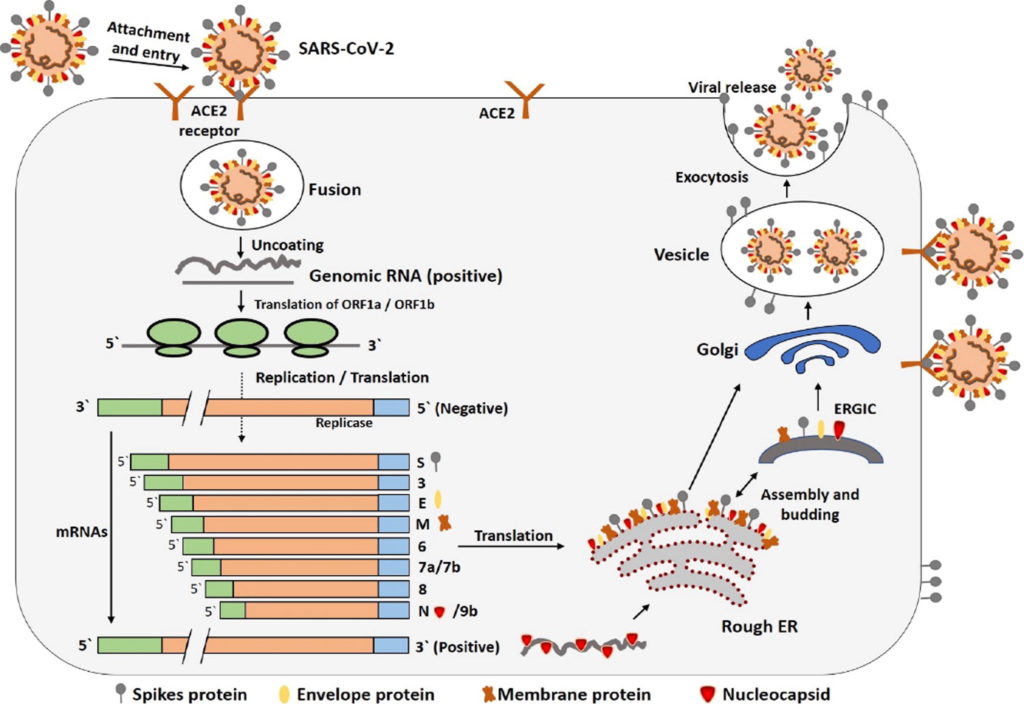
Figure 3. Life cycle of SARS-CoV-2 in host cells.
S protein binds to ACE2 receptor. After receptor binding, the conformation change in the S protein facilitates viral envelope fusion with the cell membrane through the endosomal pathway. Then SARS-CoV-2 releases RNA into the host cell. Genome RNA is translated into viral replicase polyproteins pp1a and 1ab, which are then cleaved into small products by viral proteinases. The polymerase produces a series of sub-genomic mRNAs by discontinuous transcription and finally translated into relevant viral proteins. Viral proteins and genome RNA are subsequently assembled into virions in the ER and Golgi and then transported via vesicles and released out of the cell.
Credit: https://doi.org/10.1016/j.jare.2020.03.005
Lungs are the most severely affected organ, due to the abundance of ACE 2 receptors on the type II alveolar cells. Type II alveolar cells are responsible for releasing surfactant to expand the alveolus, facilitating gas exchange at type I alveolar cells. Once entering type II alveolar cells, the viral life cycle leads to a rapid increase in the population of viruses.
Once infected by SARS-CoV-2, type II alveolar cells turn into inflammatory cells, which in turn loses its original function. This leads to the development of symptoms (cough, fever, dyspnoea) and in severe cases leading to pneumonia.
Aforementioned, there are other cells that have ACE2 receptors- this leads to a wide range of symptoms associated with digestion, taste loss, diarrhoea, fatigue, and many more.
Why is SARS-CoV-2 so contagious?
The most prominent difference between the SARSr-CoV and MERS-CoV is the type of receptor protein that the virus uses to promote the entry of the viral genome into the host cell cytoplasm. SARSr-CoV uses ACE 2 whereas MERS-CoV uses DPP4 (Dipeptidyl Peptidase 4). (See Figure 2)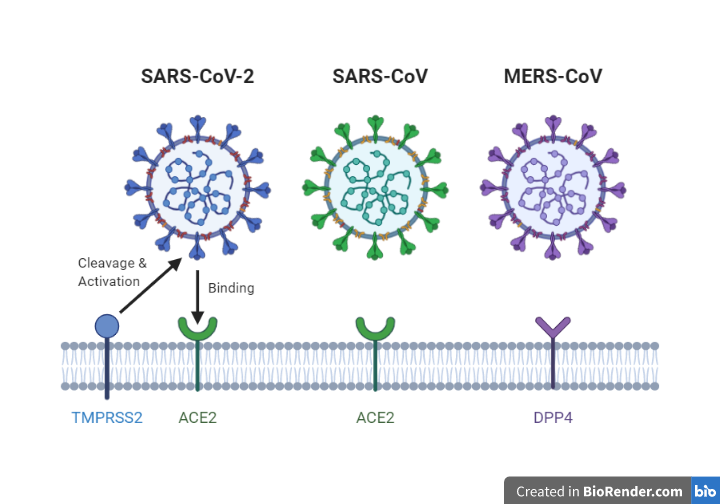
Figure 4. Overview of receptors which allows entry of the viral genome
Furthermore, there are relatively more asymptomatic cases of SARS-CoV-2 than SARS-CoV. This leads to more cases of super-spreaders than there had been in SARS-CoV and MERS-CoV.
Transmission of SARS-CoV-2
SARS-CoV-2 is mainly transmitted by respiratory droplets, which are around 5-6μm in the diameter. People can catch the virus from others through inhaling small droplets from infected people who cough or sneeze or through touching contaminated surfaces and then touching nose, mouth or eyes. It is estimated that a single droplet contains around 10-100 viruses.A single virus entering the body does not mean that one is infected; usually, one’s immune system is capable of eradicating the virus; the problem occurs when the defence line of the immune system collapses due to the entry of a large population of the virus.
The researchers also found out that the virus can survive on various types of surfaces in the absence of the host cell9. According to papers published by NIH and Princeton University, SARS-CoV-2 can survive 3 hours in aerosol; 4 hours on a copper surface; 24 hours on cardboard; 2-3days on plastics or stainless steel. This is, in general, similar to the survival time of SARS-CoV.
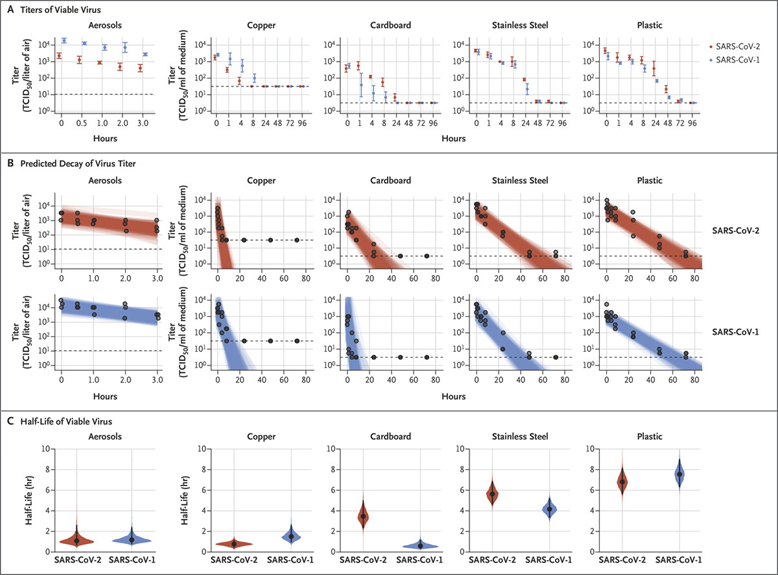
Figure 5. Viability of SARS-CoV-1 and SARS-CoV-2 in Aerosols and on Various Surfaces Credit: DOI:10.1056/NEJMc2004973
The above article was written for the purposes of general public education about the science behind the COVID-19 outbreak. It should not replace information provided by medical professionals and government officials.
References
1Masters, P. S. (2006). The Molecular Biology of Coronaviruses. Advances in Virus Research, 193–292.2 Ge, X.-Y., Li, J.-L., Yang, X.-L., Chmura, A. A., Zhu, G., Epstein, J. H., … Shi, Z.-L. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature, 503(7477), 535–538.
3,4 Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273.
5Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., … Pöhlmann, S. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell.
6Millet, J. K., & Whittaker, G. R. (2015). Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Research, 202, 120–134.
7Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An Overview of Their Replication and Pathogenesis. Coronaviruses Methods in Molecular Biology, 1–23.
8Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., … Mclellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263.
9Doremalen, N. V., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., … Munster, V. J. (2020). Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. New England Journal of Medicine.
